Circular plot of the configurations¶
This example demonstrates how to run a MM solver using K different MCMC initilization. Plot then on a circular plot all the configurations of the source localization.
# Authors: Yousra Bekhti <yousra.bekhti@gmail.com>
# Alexandre Gramfort <alexandre.gramfort@telecom-paristech.fr>
# License: BSD (3-clause)
from copy import deepcopy
import numpy as np
import mne
from mne.datasets import sample
from mne.inverse_sparse.mxne_inverse import \
(_prepare_gain, is_fixed_orient, _make_sparse_stc)
from mne.inverse_sparse.mxne_optim import norm_l2inf
from bayes_meeg.gamma_hypermodel_optimizer import (mm_mixed_norm_bayes,
compute_block_norms)
from bayes_meeg.config_plots import energy_l2half_reg, circular_brain_plot
Let us read in the fif file for MNE sample dataset corresponding to the forward, the evoked and the covariance matrix.
data_path = sample.data_path()
fwd_fname = data_path + '/MEG/sample/sample_audvis-meg-eeg-oct-6-fwd.fif'
ave_fname = data_path + '/MEG/sample/sample_audvis-ave.fif'
cov_fname = data_path + '/MEG/sample/sample_audvis-shrunk-cov.fif'
subjects_dir = data_path + '/subjects'
condition = 'Left Auditory'
# Read noise covariance matrix
noise_cov = mne.read_cov(cov_fname)
# Handling average file
evoked = mne.read_evokeds(ave_fname, condition=condition, baseline=(None, 0))
evoked.crop(tmin=0.04, tmax=0.18)
evoked = evoked.pick_types(eeg=False, meg=True)
# Handling forward solution
forward = mne.read_forward_solution(fwd_fname)
forward = mne.convert_forward_solution(forward, surf_ori=True)
Out:
365 x 365 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 59) active
Reading /home/mainak/Desktop/projects/github_repos/mne-python/examples/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left Auditory)
0 CTF compensation matrices available
nave = 55 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
Reading forward solution from /home/mainak/Desktop/projects/github_repos/mne-python/examples/MNE-sample-data/MEG/sample/sample_audvis-meg-eeg-oct-6-fwd.fif...
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Desired named matrix (kind = 3523) not available
Read MEG forward solution (7498 sources, 306 channels, free orientations)
Desired named matrix (kind = 3523) not available
Read EEG forward solution (7498 sources, 60 channels, free orientations)
MEG and EEG forward solutions combined
Source spaces transformed to the forward solution coordinate frame
Average patch normals will be employed in the rotation to the local surface coordinates....
Converting to surface-based source orientations...
[done]
Run solver method
def apply_solver(evoked, forward, noise_cov, loose=0.2, depth=0.8, K=2000):
all_ch_names = evoked.ch_names
# put the forward solution in fixed orientation if it's not already
if loose is None and not is_fixed_orient(forward):
forward = deepcopy(forward)
forward = mne.convert_forward_solution(forward, force_fixed=True)
gain, gain_info, whitener, source_weighting, mask = _prepare_gain(
forward, evoked.info, noise_cov, pca=False, depth=depth,
loose=loose, weights=None, weights_min=None)
n_locations = gain.shape[1]
sel = [all_ch_names.index(name) for name in gain_info['ch_names']]
M = evoked.data[sel]
# Whiten data
M = np.dot(whitener, M)
n_orient = 1 if is_fixed_orient(forward) else 3
# The value of lambda for which the solution will be all zero
lambda_max = norm_l2inf(np.dot(gain.T, M), n_orient)
lambda_ref = 0.1 * lambda_max
out = mm_mixed_norm_bayes(
M, gain, lambda_ref, n_orient=n_orient, K=K, return_lpp=True)
(Xs, active_sets), _, _, _, _ = out
solution_support = np.zeros((K, n_locations))
stcs, obj_fun = [], []
for k in range(K):
X = np.zeros((n_locations, Xs[k].shape[1]))
X[active_sets[k]] = Xs[k]
block_norms_new = compute_block_norms(X, n_orient)
block_norms_new = (block_norms_new > 0.05 * block_norms_new.max())
solution_support[k, :] = block_norms_new
stc = _make_sparse_stc(Xs[k], active_sets[k], forward, tmin=0.,
tstep=1. / evoked.info['sfreq'])
stcs.append(stc)
obj_fun.append(energy_l2half_reg(M, gain, stc.data, active_sets[k],
lambda_ref, n_orient))
return solution_support, stcs, obj_fun
Apply your solver
loose, depth, K = None, 0.8, 5
out = apply_solver(evoked, forward, noise_cov, loose=loose,
depth=depth, K=K)
solution_support, stcs, obj_fun = out
Out:
Average patch normals will be employed in the rotation to the local surface coordinates....
Converting to surface-based source orientations...
[done]
info["bads"] and noise_cov["bads"] do not match, excluding bad channels from both
Computing inverse operator with 305 channels.
Created an SSP operator (subspace dimension = 3)
estimated rank (mag + grad): 302
Setting small MEG eigenvalues to zero.
Not doing PCA for MEG.
Total rank is 302
Whitening lead field matrix.
Running iter 0
Running iter 1
iter 0 - active set 7498
iter 1 - active set 85
iter 2 - active set 10
iter 3 - active set 6
iter 4 - active set 4
iter 5 - active set 4
iter 6 - active set 4
iter 7 - active set 4
iter 8 - active set 4
iter 9 - active set 4
sample 0 - lppSamp -402881.936738 - relResSamp 2.17437229397 - lppMAP -1271.35913719 - relResSamp 0.709948257298
Running iter 0
Running iter 1
iter 0 - active set 7498
iter 1 - active set 91
iter 2 - active set 11
iter 3 - active set 8
iter 4 - active set 6
iter 5 - active set 5
iter 6 - active set 5
iter 7 - active set 4
iter 8 - active set 4
iter 9 - active set 4
sample 1 - lppSamp -486045.920556 - relResSamp 2.32125847403 - lppMAP -1275.42865313 - relResSamp 0.710445665549
Running iter 0
Running iter 1
iter 0 - active set 7498
iter 1 - active set 131
iter 2 - active set 7
iter 3 - active set 5
iter 4 - active set 5
iter 5 - active set 5
iter 6 - active set 5
iter 7 - active set 5
iter 8 - active set 5
iter 9 - active set 5
sample 2 - lppSamp -538217.425309 - relResSamp 2.37773935702 - lppMAP -1273.21837565 - relResSamp 0.699096440769
Running iter 0
Running iter 1
iter 0 - active set 7498
iter 1 - active set 118
iter 2 - active set 11
iter 3 - active set 8
iter 4 - active set 6
iter 5 - active set 6
iter 6 - active set 5
iter 7 - active set 5
iter 8 - active set 5
iter 9 - active set 5
sample 3 - lppSamp -576993.083229 - relResSamp 2.46439589067 - lppMAP -1302.82192397 - relResSamp 0.713972774165
Running iter 0
Running iter 1
iter 0 - active set 7498
iter 1 - active set 120
iter 2 - active set 9
iter 3 - active set 7
iter 4 - active set 7
iter 5 - active set 6
iter 6 - active set 6
iter 7 - active set 6
iter 8 - active set 6
iter 9 - active set 6
sample 4 - lppSamp -607331.969873 - relResSamp 2.45626883325 - lppMAP -1297.15703901 - relResSamp 0.697444448733
Plotting of all configurations intro a circular plot
circular_brain_plot(forward, solution_support, stcs, obj_fun)
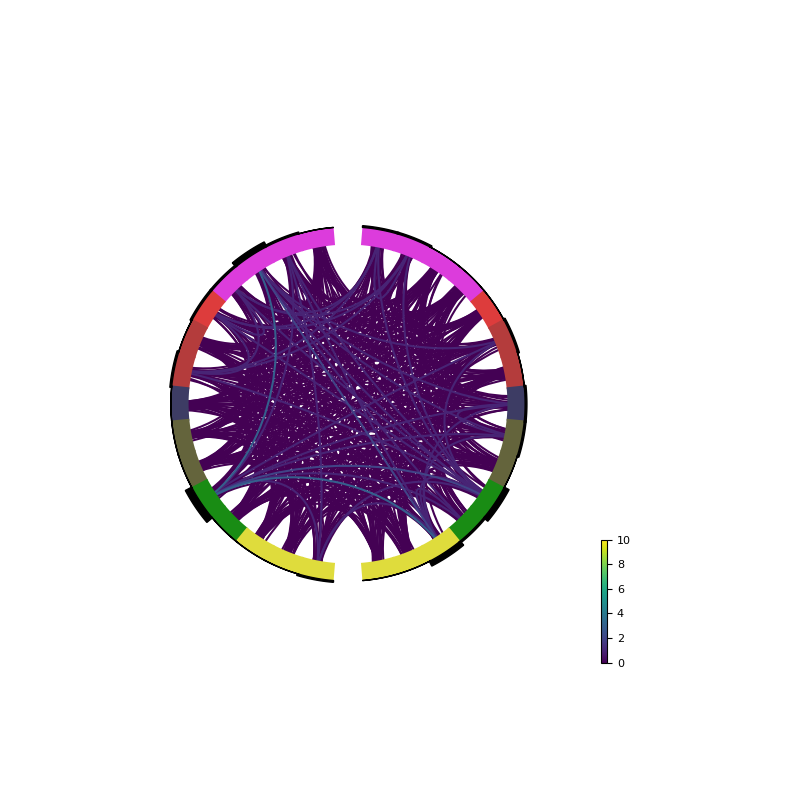
Out:
Reading labels from parcellation...
read 75 labels from /home/mainak/Desktop/projects/github_repos/mne-python/examples/MNE-sample-data/subjects/sample/label/lh.aparc.a2009s.annot
read 75 labels from /home/mainak/Desktop/projects/github_repos/mne-python/examples/MNE-sample-data/subjects/sample/label/rh.aparc.a2009s.annot
Total running time of the script: ( 17 minutes 58.544 seconds)